 v.231201
v.231201
About
The SysteMHC Atlas project is part of the Human Immuno-Peptidome Project (HIPP). HIPP was launched in 2015 as a new initiative of the Human Proteome Organization (HUPO). The long-term goal of the HIPP is to map the entire repertoire of peptides presented by MHC molecules using mass spectrometry technologies and make its robust analysis accessible to any immunologist. Toward this end, HIPP plans to embrace partnerships, prioritize technology development and maximize data sharing. The SysteMHC Atlas project supports this latter goal and this is an updated one (Second Version) .
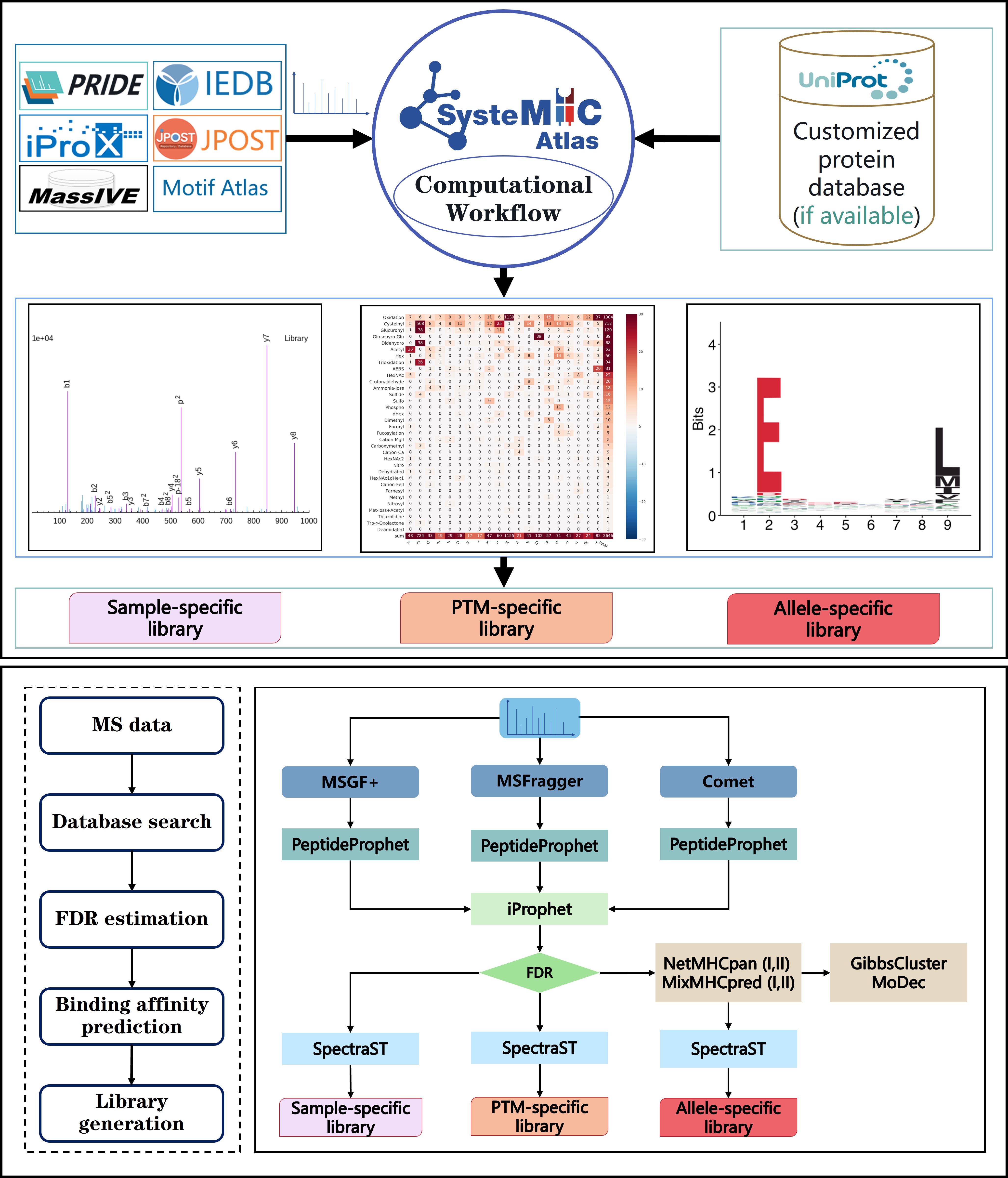
Overview of the updated SysteMHC build process:
Following are the key parameters:
| General Parameters | |
|---|---|
| Parameter | Value |
| Precursor tolerance | 10 ppm |
| High accuracy fragment ion tolerance (Orbi/TOF/QE) | 10 ppm |
| Low accuracy fragment ion tolerance | 0.5Da |
| Digestion specificity | unspecific |
| Open Search Parameters | ||
|---|---|---|
| Delta mass | Name | Description |
| -105.0248 | Met-loss+acetaldehyde | Met-loss+acetaldehyde |
| -89.0299 | Met-loss+Acetyl | Met-loss+Acetylation |
| -33.9877 | Cys->Dha | Dehydroalanine |
| -32.0085 | Met->AspSA | Methionine-oxidation-to-aspartic-semialdehyde |
| -30.0106 | Pro->Pyrrolidinone | Proline-oxidation-to-pyrrolidinone |
| -29.9928 | Met->Hse | Homoserine |
| -18.0106 | Glu->pyro-Glu | Glu->pyro-Glu |
| -17.0265 | Gln->pyro-Glu | Gln->pyro-Glu |
| -2.0157 | Didehydro | second-amino-3-oxo-butanoic_acid |
| -1.9979 | Met->Glu | Met->Glu substitution |
| -1.9615 | Cys->Thr | Cys->Thr substitution |
| -0.984 | Amidation | Amidation |
| +0.984 | Deamidated | Deamidation |
| +3.9949 | Trp->Kynurenin | tryptophan-oxidation-to-kynurenin |
| +12.0 | Thiazolidine | formaldehyde-adduct |
| +13.9793 | Trp->Oxolactone | Tryptophan-oxidation-to-oxolactone |
| +14.0157 | Methyl | Methylation |
| +15.9949 | Oxidation | Oxidation or Hydroxylation |
| +19.9898 | Trp->Hydroxykynurenin | tryptophan-oxidation-to-hydroxykynurenin |
| +21.9694 | Cation-Mg | Replacement-of-2-protons-by-magnesium |
| +21.9819 | Cation-Na | Sodium-adduct |
| +23.9581 | Cation-Al | Replacement-of-3-protons-by-aluminium |
| +26.0157 | Delta-H(2)C(2) | Acetaldehyde+26 |
| +27.9949 | Formyl | Formylation |
| +28.0313 | Dimethyl | di-Methylation |
| +28.9902 | Nitrosyl | nitrosylation |
| +29.9742 | quinone | Cquinone |
| +30.0106 | hydroxymethyl | hydroxymethyl |
| +31.9721 | persulfide | persulfide |
| +31.9898 | dihydroxy | dihydroxy |
| +37.9469 | Cation-Ca | Replacement-of-2-protons-by-calcium |
| +37.9558 | Cation-K | Replacement-of-proton-by-potassium |
| +42.0106 | Acetyl | Acetylation |
| +42.047 | Trimethyl | tri-Methylation |
| +43.0058 | Carbamyl | Carbamylation |
| +43.9898 | Carboxy | Carboxylation |
| +44.9851 | Nitro | Oxidation-to-nitro |
| +47.9847 | Trioxidation | cysteine-oxidation-to-cysteic-acid |
| +53.9193 | Cation-Fe | Replacement of 2 protons by iron |
| +58.0055 | Carboxymethyl | Iodoacetic acid derivative |
| +68.0262 | Crotonyl | Crotonylation |
| +70.0418 | Crotonaldehyde | Crotonaldehyde |
| +79.9568 | Sulfo | O-Sulfonation |
| +79.9663 | Phospho | Phosphorylation |
| +80.0374 | Gly->His | Gly->His |
| +86.0004 | Malonylation | Malonylation |
| +100.016 | Succinyl | Succinic-anhydride-labeling-reagent-light-form(N-term&K) |
| +119.0041 | Cysteinyl | Cysteinylation |
| +119.0371 | pyridylacetyl | pyridylacetyl |
| +146.0579 | Fucosylation | Fucose |
| +162.0528 | Hex | Hexose |
| +176.0321 | Glucuronyl | hexuronic acid |
| +178.0477 | Gluconoylation | Gluconoylation |
| +183.0354 | AEBS | Aminoethylbenzenesulfonylation |
| +203.0794 | HexNAc | N-Acetylhexosamine |
| +204.1878 | Farnesylation | Farnesylation |
| +210.1984 | Myristoylation | Myristoylation |
| +229.014 | Pyridoxal Phosphate | Pyridoxal phosphate |
| +238.2297 | Palmitoylation | Palmitoylation |
| +340.1006 | Glucosylgalactosyl | glucosylgalactosyl hydroxylysine |
| +349.1373 | HexNAc1dHex1 | HexNAc1dHex1 |
| +365.1322 | Hex1HexNAc1 | Hex1HexNAc1 |
| +406.1587 | HexNAc2 | HexNAc2 |
| +541.0611 | ADP-Ribosyl | ADP Ribose addition |
In general, the computational pipeline was based on Trans-proteomic Pipeline version 6.0.0 -- We used msConvert to convert mass spectrometric raw files into mzML/mzXML files with default settings. And three search engines (i.e. Comet, MSFragger and MSGF+) were used to perform (closed/offset) database searches with a target-decoy strategy. The instrument-specific search parameters were described above. For statistical validation, PeptideProphet was first applied with the accurate mass model enabled. Then, iProphet was used to combine the outputs of PeptideProphet from all three search engines.
To predict the binding affinity, NetMHCpan-4.1 and MixMHCpred-2.2 were used for MHC Class I peptides with the default settings, and NetMHCIIpan-4.1 and MixMHC2pred-2.0 were used for MHC class II peptides. The Motifs of predicted binders of Class I and II were deconvoluted by GibbsCluster-2.0 and MoDec-1.2, respectively. Additionally, we used MixMHCp-2.1 and MoDec-1.2 to perform direct motif deconvolution for the MHC class I immunopeptidome and MHC class II immunopeptidome, respectively.
The Annotation score for the peptides annotated for the allele is calculated using the strategy described in Caron et al. eLife 2015, based on the binding affinity prediction by NetMHCpan and NetMHCIIpan. Specificlly, it is calculated by dividing the second lowest IC50 value (second best predicted allele) by the lowest IC50 value (best predicted allele).
At a 1% peptide level FDR estimated by iProphet, spectral libraries were constructed by SpectraST with default settings for consensus library building. For each sample, a sample-specific consensus spectral library was generated. For each allele, an allele-specific spectral library was generated on the atlas level that contains consensus spectra of peptides from different samples ranging from diverse tissue types and disease types. In the allele-specific spectral library, the same peptide ions generated under various fragmentation methods were specified and kept separated as different library entries. For each PTM, a PTM-specific spectral library was generated.
The current build version is 230601.
How to cite SysteMHC Atlas
Please acknowledge the SysteMHC Atlas in your publications by citing the following manuscript:
The computational pipeline
The computational pipeline SysteMHC-pipeline is provided by GitHub repository.
The team behind SysteMHC Atlas:
- Wenguang Shao (Professor, PhD)
- Xiaoxiang Huang (PhD candidate)
- Ziao Gan (Undergraduate student)
- Haowei Cui (Undergraduate student)
- Tian Lan (Graduated student)
Licenses
SysteMHC Atlas is made available under the Open Database License: http://opendatacommons.org/licenses/odbl/1.0/.
Any rights in individual contents of the database are licensed under the Database Contents License: http://opendatacommons.org/licenses/odbl/1.0/.
Contact us:
- Wenguang Shao, ✉ wshao@sjtu.edu.cn
- Xiaoxiang Huang, ✉ xx_huang@sjtu.edu.cn